
The Cytoscape Attribute Browser provides a tabular view of data that have been loaded into Cytoscape and attached to nodes, edges or networks. 1C) to further enhance network visualization. 1A and B), images enable shading, highlights and other aesthetic effects to be applied to nodes ( Fig. In addition to rich, multivariate visualizations ( Fig. Placement and sizing of images can also be directly controlled within Cytoscape. Images can be mapped to nodes using Cytoscape's VizMapper framework, which connects the visualization properties of the network to attribute data. Given a URL, Cytoscape will try to read that URL and generate an image for display. For cases involving large numbers of custom images, Cytoscape also allows images to be loaded as node attributes by providing a Uniform Resource Locator (URL) address for each image. Using this new feature, images are loaded using a standard file browser or by dragging and dropping them into a pool of available images maintained by Cytoscape. Version 2.8 introduces the ability for non-programmers to specify images through the Cytoscape GUI and to map these images to nodes using Cytoscape's standard VizMapper interface. To represent multivariate data associated with a node, Cytoscape can control the visualization of each node using a custom graphical image (since version 2.3) through a programming API. Scalar data can be linked to simple visualization properties such as node color, shape or size as node and edge attributes. Taken together, however, these features provide a mechanism for expressing relationships between sets of data while simultaneously visualizing the integrated results.Ī key function of Cytoscape is to allow diverse types of attribute data to be visualized on the nodes and edges of a biological network. Separately, each of these features provides useful new capabilities to Cytoscape. The second feature is the introduction of spreadsheet-like equations into Cytoscape's Attribute Browser to enable advanced transformation and combination of datasets directly within Cytoscape.
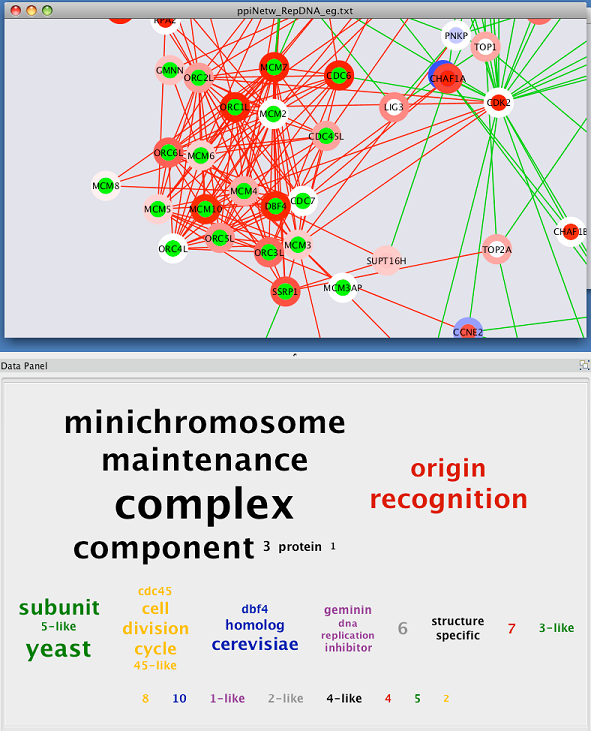
The first feature allows non-programmers to map graphical images onto nodes, which greatly increases the power and flexibility with which integrated data can be visualized. Version 2.8 of Cytoscape has introduced two significant new features that improve its ability to integrate and visualize complex datasets. shape, color, size) as well as to perform complex network searches, filtering operations and other analysis. Attribute values can be used to control visual aspects of nodes and edges (e.g. Data are integrated with the network using attributes, which map nodes or edges to specific data values such as gene expression levels or protein functions. genes, proteins, cells, patients) represented as nodes and biological interactions represented as edges between nodes. Its central organizing principle is a network graph, with biological entities (e.g.

Thus data integration, along with integrated visualization, is a key.Ĭytoscape, now in its eighth year of development, has become a standard tool for integrated analysis and visualization of biological networks ( Cline et al., 2007 Shannon et al., 2003). Although various datasets can appear quite different in quality and quantity, they all are reflections of the same underlying biological system and its responses.
These network data extend and compliment a great deal of other information available in the biomedical sciences. Vast datasets are being gathered to delineate networks of various types and levels, including networks of genetic and protein–protein interaction networks of transcriptional and post-transcriptional regulation, social networks and multiscale networks that span several of these domains ( Fowler et al., 2009 Tan et al., 2007).


 0 kommentar(er)
0 kommentar(er)
